The growth of microbes must consume nutrients (substrates) and transform those nutrients into products, conserving energy from the transformation. These transformations, to date, must be atomically and electronically conserved, resulting in overall reaction stoichiometries of substrate and products involved for energy conservation (catabolic reactions) and biomass production (anabolic reactions). Across the tree of life, macromolecule atomic and electronic (degree of reduction) compositions are fairly conserved. Therefore, to constrain the contribution of component populations in a microbial community, the catabolic reactions provide a primary target for quantification.
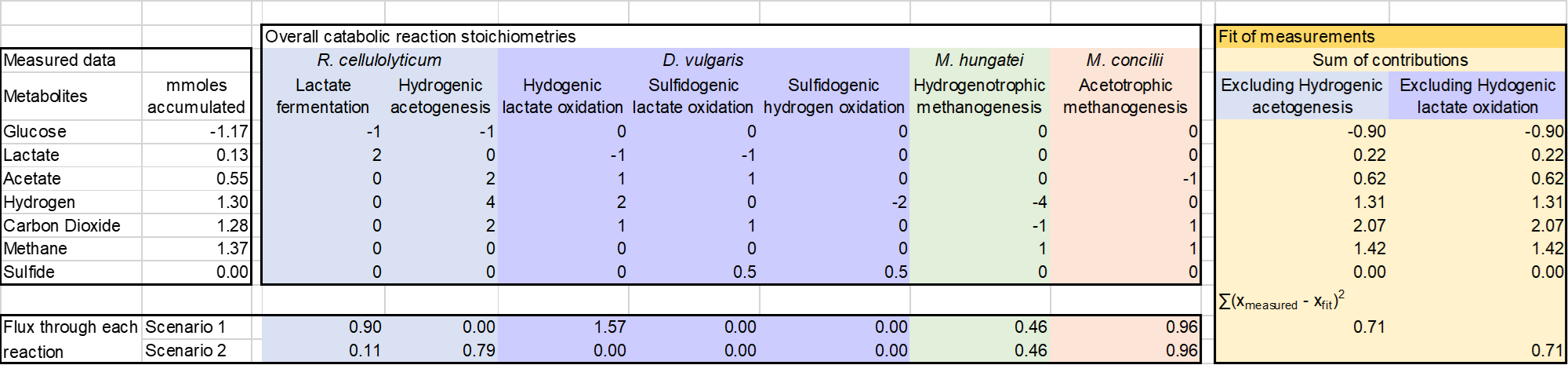
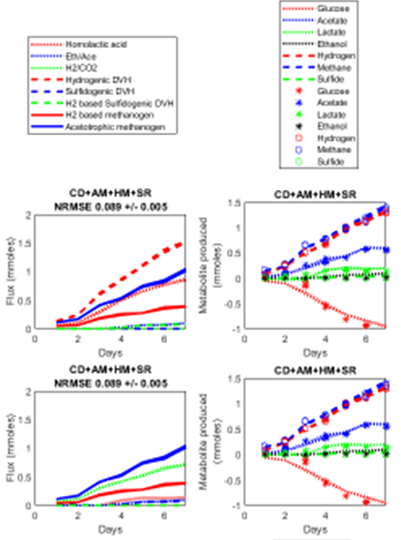
A set of overall reactions representing the documented metabolisms of the component populations were fit into the daily measured amounts of metabolites in the constructed synthetic communities. The metabolism of R. cellulolyticum was modeled to consume glucose to eliminate artifacts from cellulase production. The overall metabolism of R. cellulolyticum was modeled as lactate fermentation, hydrogenic acetogenesis. The low observed ethanol in ethanol/acetate fermentations, accounting for less than 0.36% of the carbon flux, led us to remove this pathway from analysis. D. vulgaris was modeled to oxidize lactate to acetate, coupled to reduction of protons or sulfate, or oxidize H2 coupled to sulfate reduction. The methanogens were represented by hydrogenotrophic methanogenesis and acetoclastic methanogenesis. The resulting fits indicate the relative contribution of each microorganism to the overall observed transformations and provide a basis for quantifying metabolic contributions. Deviations between the inferred metabolic transformations and the measured transformations indicate uncertainty in our understanding of the metabolic transformations or limitations in analytical capacity. To access redundancy of metabolic pathways in this stoichiometric model, each reaction pathway was sequentially excluded before fitting and the impact on model fit was evaluated. These simulations identified two and three possible flux distributions through the quad-culture in the control and sulfate addition conditions with equal goodness of fit to the experimental data.
-
The general concept of the stoichiometric model under control condition
-
In the control condition, two possible flux distributions show the equal goodness of the fitting
-
reaction matrix and metadata measured from experiment are as the input of the model. The matrix and metadata in this paper were put in the script&data folder.
-
put the csv files of reaction matrix, metadata and matlab script in one folder, and run the script.
We only used GC analysis the CO2 in the gas phase, but it was not a direct representation of CO2 produced from the reactions because of the buffer and pH effects on equilibrium, so when did fitting, we remove the CO2 from the matrix.
Kristopher A. Hunt hunt0362@uw.edu
Dongyu Wang dywang@ou.edu
Chongle Pan cpan@ou.edu